Recent genomic studies in adult and pediatric acute myeloid leukemia (AML) demonstrated recurrent in-frame tandem duplications (TD) in exon 13 of upstream binding transcription factor ( UBTF) (PMID: 35176137, PMID:37085611). UBTF-TDs are subtype-defining genomic alterations associated with poor outcomes that account for ~4.3% of AMLs in childhood and up to 3% in adult AMLs under the age of 60. UBTF-TD AMLs are characterized by HOXA/HOXB dysregulation, trisomy 8 or normal cytogenetics and frequent FLT3-ITD and/or WT1 mutations. A more comprehensive investigation into the clinicopathological features of UBTF-TD AMLs is needed to better understand disease features for appropriate diagnosis and to develop more effective therapies.
Here, we present 94 unique pediatric cases harboring a tandem duplication in exon 13 of UBTF with a median size of 60bp (range 21-1173bp). Of these 94 cases, 6 (6.4%) were diagnosed with childhood MDS with increased blasts (WHO), suggesting that like in adults, UBTF-TDs are not only confined to AML. UBTF-TD AMLs are associated with an overall lower median white blood cell (WBC) when compared to other common HOXA/B dysregulated leukemias ( UBTF-TD median = 10.2x10 9/L, NPM1-mutation median = 21.8x10 9/L, NUP98-rearranged ( NUP98r) median = 97x10 9/L). Available immunophenotyping on 32 U BTF-TD tumors demonstrated positivity for HLA-DR (31/31; 100%), CD117 (28/29; 95%), and CD34 (26/30; 86%) with variable positivity for CD7 (17/30; 56%) and CD64 (13/25; 52%). A limited number of cases tested for CD123 (n=11) and CD11c (n=15) were all positive for the respective marker. Furthermore, morphologic assessment of these cases revealed pleomorphic blasts with occasional Auer rods and increased immature myeloid cells with salmon-colored granules, commonly with background multilineage dysplasia and increased erythroid precursors.
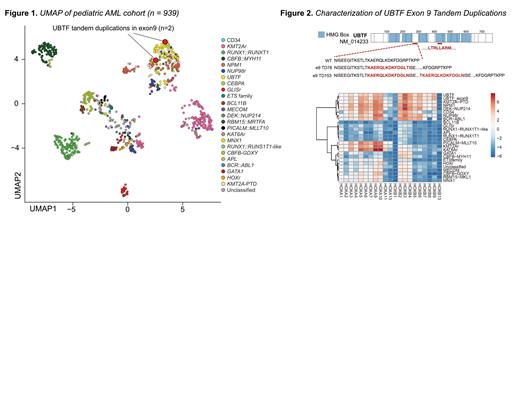
At the transcriptional level, UBTF-TD leukemias are largely similar to NPM1-mutated and NUP98::NSD1 AMLs with the exception of UBTF-TD cases having marked increase in expression of histone genes (e.g., HIST1H4F and HIST1H2B1) and a subset of posterior HOXB cluster genes (e.g., HOXB9). Further inspection using UMAP (Uniform Manifold Approximation and Projection) of expression profiles across different pediatric AML cohorts revealed two unclassified cases of AML that clustered with the UBTF-TD subtype ( Figure 1). One case had a WT1p.S364* and the other a somatic GATA2 p.R338fs and FLT3-ITD, but did not contain an exon 13 duplication or other defining alterations. Upon closer inspection of the UBTF gene in these cases, we uncovered that each case had an in-frame tandem duplication in exon 9 of UBTF encoding a region between HMG box 2 and 3 with features similar to those in exon 13 ( Figure 2). We therefore postulated that exon 9 TDs ( UBTF-e9) could represent a functionally equivalent subgroup of UBTF-TDalterations.
To test this hypothesis, cord-blood CD34+ (cbCD34+) cells were transduced to test the transforming potential of UBTF-e9 duplications. Like with UBTF-e13 duplications, cbCD34+ cells expressing UBTF-e9 duplications displayed increased proliferation compared to UBTF -WT and lentiviral empty vector control in liquid culture; and increased colony counts and replating capacity, increased CD117 expression, and a more blast-like morphology in CFU assays. Collectively, these data suggest that like TDs in exon 13, exon 9 TDs are also sufficient to promote leukemogenic phenotypes.
In conclusion, we find that UBTF-TDs occur in both MDS and AML and are associated with lower WBC count, myelodysplasia, and stem cell-related surface marker expression. We have also expanded the spectrum of UBTFalterations in myeloid neoplasms through the identification of rare UBTF duplications in exon 9 that induce a similar transcriptional signature to exon 13 UBTF-TDs. The recent identification of UBTF-TDs in adult and pediatric high-risk myeloid neoplasms has highlighted the importance of understanding and appropriately diagnosing tumors with this genomic alteration, including a new class of alterations that will not be detected by PCR-based screening strategies that specifically target exon 13 of UBTF.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal